A theoretical 3D structure of GDH4 from Ref was generated with the homology-modeling program ESyPred3D using as the template the structure of bovine GDH3 (PDB # 1HWZ).
The modelisation and the drawing of a putative structure of GDH4 was performed with the protein structure homology-modeling program DeepView (SwissPdb-Viewer).
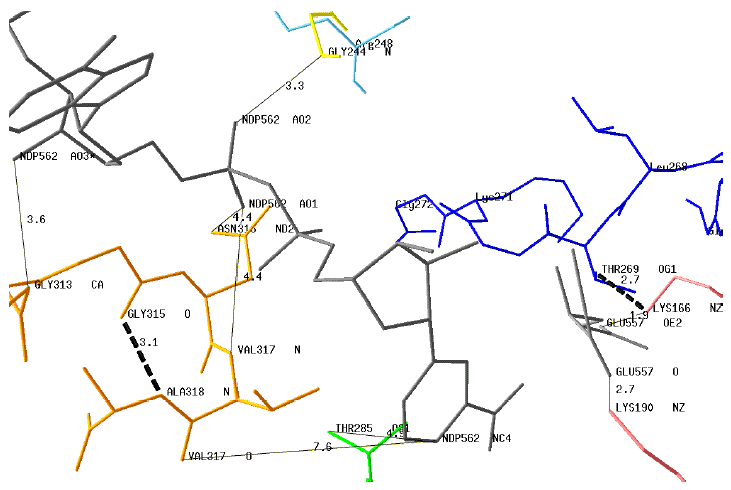
1. Some interactions (plain lines in figure below) between the motif G313AGNVA318 or key residues and the coenzyme are indicated :
- NDP562AO3 - Gly313CA
- NDP562AO1 - Asn316ND2
- NDP562AO1 - Val317N
- NDP562AO2 - Gly244N
- NDP562NC4 - Thr285OG1
2. The distances between the protonated carbon atom of the nicotinamide moiety (NDP562NC4) are too long for direct interactions with the motif G313AGNVA318.
3. However, this motif is stabilized by an internal H-bond Gly315O - Ala318N (dotted line).
4. Two distances (Glu557OE2 - Lys166NZ and Glu557O - Lys190NZ) are compatible with H-bond interactions between the enzyme and Glu.
5. The position of the motif G266VLTGKG272 is shown with the potential H-bond Lys166NZ - Thr269OG1.